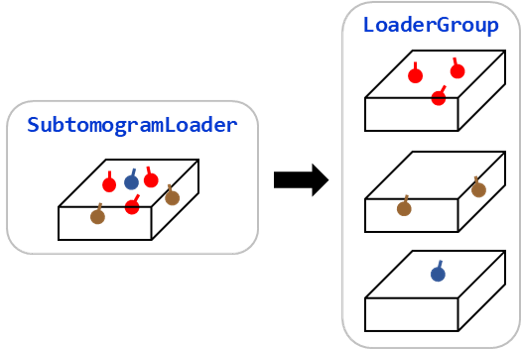
Loader Group
The groupby() of subtomogram loaders returns a LoaderGroup object.
An LoaderGroup object is very similar to those returned by groupby()
methods of polars.DataFrame, Molecules or pandas.DataFrame.
from acryo import SubtomogramLoader
loader = SubtomogramLoader(image, molecules)
for cluster, ldr in loader.groupby("cluster_id"):
assert (out.molecules.features["cluster_id"] == cluster).all()
LoaderGroup has many methods of the same name as those in SubtomogramLoader.
Group-wise Averaging
LoaderGroup supports all the averaging methods.
average()average_split()
In LoaderGroup version, result is returned as a dict
of group key and the averages.
Group-wise Alignment
LoaderGroup also supports all the alignment methods
align()align_no_template()align_multi_templates()
In LoaderGroup version, result is returned as an updated LoaderGroup.
If you want to collect aligned Molecules objects, following codes are
essentially equivalent.
# call align() for each loader
aligned = []
for cluster, ldr in loader.groupby("cluster_id"):
out = ldr.align(template)
aligned.append(out.molecules)
# call align() of the LoaderGroup object.
aligned = []
for cluster, ldr in loader.groupby("cluster_id").align(template):
aligned.append(out.molecules)
Since each group does not necessarily composed of the same molecules, you can use a mapping of templates for alignment functions.
templates = {
0: template0,
1: template1,
2: template2,
}
aligned = loader.groupby("cluster_id").align_multi_templates(templates)